import mathimport numpy as npimport matplotlib.pyplot as pltimport pandas as pdimport torchimport GPyimport jaximport gpytorchimport botorchimport tinygpimport jax.numpy as jnpimport optaxfrom IPython.display import clear_outputfrom sklearn.preprocessing import StandardScaler
WARNING:absl:No GPU/TPU found, falling back to CPU. (Set TF_CPP_MIN_LOG_LEVEL=0 and rerun for more info.)
Data
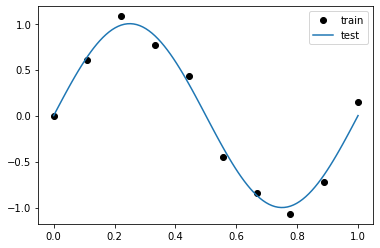
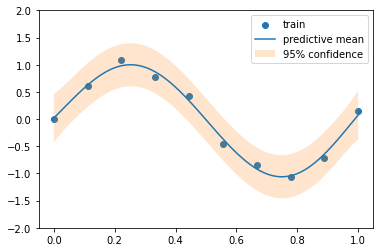
0 ) # We don't want surprices in a presentation :) = 10 = torch.linspace(0 , 1 , N)= torch.sin(train_x * (2 * math.pi)) + torch.normal(0 , 0.1 , size= (N,))= torch.linspace(0 , 1 , N* 10 )= torch.sin(test_x * (2 * math.pi))
'ko' , label= 'train' ); = 'test' ); ;
Defining kernel
\[\begin{equation}
\sigma_f^2 = \text{variance}\\
\ell = \text{lengthscale}\\
k_{RBF}(x_1, x_2) = \sigma_f^2 \exp \left[-\frac{\lVert x_1 - x_2 \rVert^2}{2\ell^2}\right]
\end{equation}\]
GPy
= GPy.kern.RBF(input_dim= 1 , variance= 1. , lengthscale= 1. )
rbf. value constraints priors
variance
1.0
+ve
lengthscale
1.0
+ve
GPyTorch
= gpytorch.kernels.ScaleKernel(gpytorch.kernels.RBFKernel())= 1. # variance = 1. # lengthscale
ScaleKernel(
(base_kernel): RBFKernel(
(raw_lengthscale_constraint): Positive()
)
(raw_outputscale_constraint): Positive()
)
TinyGP
def RBFKernel(variance, lengthscale):return jnp.exp(variance) * tinygp.kernels.ExpSquared(scale= jnp.exp(lengthscale))= RBFKernel(variance= 1. , lengthscale= 1. )
<tinygp.kernels.Product at 0x7f544039d710>
Define model
\[
\sigma_n^2 = \text{noise variance}
\]
GPy
= GPy.models.GPRegression(train_x.numpy()[:,None ], train_y.numpy()[:,None ], gpy_kernel)= 0.1
Model : GP regressionObjective : 16.757933772959404Number of Parameters : 3Number of Optimization Parameters : 3Updates : True
GP_regression. value constraints priors
rbf.variance
1.0
+ve
rbf.lengthscale
1.0
+ve
Gaussian_noise.variance
0.1
+ve
GPyTorch
class ExactGPModel(gpytorch.models.ExactGP):def __init__ (self , train_x, train_y, likelihood, kernel):super ().__init__ (train_x, train_y, likelihood)self .mean_module = gpytorch.means.ConstantMean()self .covar_module = kerneldef forward(self , x):= self .mean_module(x)= self .covar_module(x)return gpytorch.distributions.MultivariateNormal(mean_x, covar_x)= gpytorch.likelihoods.GaussianLikelihood()= ExactGPModel(train_x, train_y, gpytorch_likelihood, gpytorch_kernel)= 0.1
ExactGPModel(
(likelihood): GaussianLikelihood(
(noise_covar): HomoskedasticNoise(
(raw_noise_constraint): GreaterThan(1.000E-04)
)
)
(mean_module): ConstantMean()
(covar_module): ScaleKernel(
(base_kernel): RBFKernel(
(raw_lengthscale_constraint): Positive()
)
(raw_outputscale_constraint): Positive()
)
)
TinyGP
def build_gp(theta, X):= theta[0 ] = jnp.exp(theta[1 :])= variance * tinygp.kernels.ExpSquared(lengthscale)return tinygp.GaussianProcess(kernel, X, diag= noise_variance, mean= mean)= build_gp(theta= np.array([0. , 1. , 1. , 0.1 ]), X= train_x.numpy())# __repr__
<tinygp.gp.GaussianProcess at 0x7f5440401850>
Train the model
GPy
= 50 )
Model : GP regressionObjective : 3.944394423452163Number of Parameters : 3Number of Optimization Parameters : 3Updates : True
GP_regression. value constraints priors
rbf.variance
0.9376905183253631
+ve
rbf.lengthscale
0.2559000163858406
+ve
Gaussian_noise.variance
0.012506184441481319
+ve
GPyTorch
= gpytorch.mlls.ExactMarginalLogLikelihood(gpytorch_likelihood, gpytorch_model)# Mean # Variance # Lengthscale # Noise variance
/opt/conda/lib/python3.7/site-packages/botorch/fit.py:143: UserWarning:CUDA initialization: CUDA unknown error - this may be due to an incorrectly set up environment, e.g. changing env variable CUDA_VISIBLE_DEVICES after program start. Setting the available devices to be zero. (Triggered internally at /opt/conda/conda-bld/pytorch_1634272168290/work/c10/cuda/CUDAFunctions.cpp:112.)
Parameter containing:
tensor([0.0923], requires_grad=True)
tensor(0.9394, grad_fn=<SoftplusBackward0>)
tensor([[0.2560]], grad_fn=<SoftplusBackward0>)
tensor([0.0124], grad_fn=<AddBackward0>)
TinyGP
from scipy.optimize import minimizedef neg_log_likelihood(theta, X, y):= build_gp(theta, X)return - gp.condition(y)= jax.jit(jax.value_and_grad(neg_log_likelihood))= minimize(obj, [0. , 1. , 1. , 0.1 ], jac= True , args= (train_x.numpy(), train_y.numpy()))0 ], np.exp(result.x[1 :])
(0.09213499552879165, array([0.9395271 , 0.25604163, 0.01243025]))
Inference
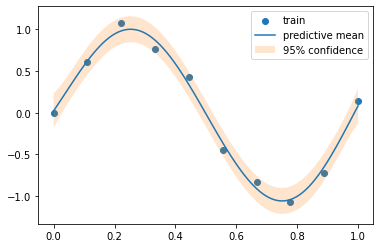
def plot_gp(pred_y, var_y):= var_y ** 0.5 = 'train' )= 'predictive mean' )- 2 * std_y.ravel(), + 2 * std_y.ravel(), alpha= 0.2 , label= '95 % c onfidence' )
GPy
= gpy_model.predict(test_x.numpy()[:, None ])
GPyTorch
eval ()with torch.no_grad(), gpytorch.settings.fast_pred_var():= gpytorch_likelihood(gpytorch_model(test_x))= pred_dist.mean, pred_dist.variance
TinyGP
= build_gp(result.x, train_x.numpy())= tinygp_model.predict(train_y.numpy(), test_x.numpy(), return_var= True )
Tiny GP on CO2 dataset
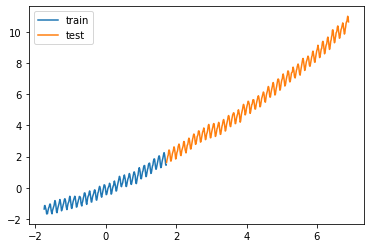
= pd.read_csv("data/co2.csv" )# Train test split = data["0" ].iloc[:290 ].values.reshape(- 1 , 1 )= data["0" ].iloc[290 :].values.reshape(- 1 , 1 )= data["1" ].iloc[:290 ].values= data["1" ].iloc[290 :].values# Scaling the dataset = StandardScaler()= Xscaler.fit_transform(X)= Xscaler.transform(X_test)= StandardScaler()= yscaler.fit_transform(y.reshape(- 1 , 1 )).ravel()= yscaler.transform(y_test.reshape(- 1 , 1 )).ravel()
= 'train' ); = 'test' ); ;
class SpectralMixture(tinygp.kernels.Kernel):def __init__ (self , weight, scale, freq):self .weight = jnp.atleast_1d(weight)self .scale = jnp.atleast_1d(scale)self .freq = jnp.atleast_1d(freq)def evaluate(self , X1, X2):= jnp.atleast_1d(jnp.abs (X1 - X2))[..., None ]return jnp.sum (self .weight* jnp.prod(- 2 * jnp.pi ** 2 * tau ** 2 / self .scale ** 2 )* jnp.cos(2 * jnp.pi * self .freq * tau),=- 1 ,def build_spectral_gp(theta):= SpectralMixture("log_weight" ]),"log_scale" ]),"log_freq" ]),return tinygp.GaussianProcess(= jnp.exp(theta["log_diag" ]), mean= theta["mean" ]
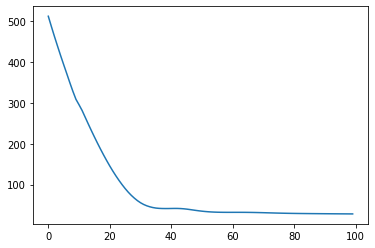
= 4 # Number of mixtures = 0.4 1 )= {"log_weight" : np.abs (np.random.rand(K))/ div_factor,"log_scale" : np.abs (np.random.rand(K))/ div_factor,"log_freq" : np.abs (np.random.rand(K))/ div_factor,"log_diag" : np.abs (np.random.rand(1 ))/ div_factor,"mean" : 0. ,@jax.jit @jax.value_and_grad def loss(theta):return - build_spectral_gp(theta).condition(y)# opt = optax.sgd(learning_rate=0.001) = optax.adam(learning_rate= 0.1 )= opt.init(params)= []for i in range (100 ):= loss(params)= opt.update(grads, opt_state)= optax.apply_updates(params, updates)= True )print (f"iter { i} , loss { loss_val} " )= build_spectral_gp(params)
iter 99, loss 27.987701416015625
{'log_diag': DeviceArray([-2.7388687], dtype=float32),
'log_freq': DeviceArray([-3.6072493, -3.1795945, -3.4490397, -2.373117 ], dtype=float32),
'log_scale': DeviceArray([3.9890492, 3.8530042, 4.0878096, 4.4860597], dtype=float32),
'log_weight': DeviceArray([-1.3715047, -0.6132469, -2.413771 , -1.6582283], dtype=float32),
'mean': DeviceArray(0.38844627, dtype=float32)}
= opt_gp.predict(y, X_test, return_var= True )= 'k' )+ np.sqrt(var), mu - np.sqrt(var), color= "C0" , alpha= 0.5 = "C0" , lw= 2 )# plt.xlim(t.min(), 2025) "year" )= plt.ylabel("CO$_2$ in ppm" )
Idea
\(K_\mathbf{y} = \text{cov_function}(X_{train}, X_{train}, \sigma_f, \ell, \sigma_n)\)
GP Loss: \(\log p(\mathbf{y} \mid \mathbf{X}, \theta)=-\frac{1}{2} \mathbf{y}^{T} K_{y}^{-1} \mathbf{y}-\frac{1}{2} \log \left|K_{y}\right|-\frac{n}{2} \log 2 \pi\)
Minimize inverse term fully
Now, Minimize both togather
Appendix
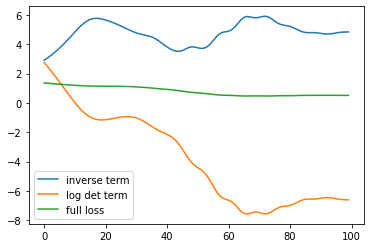
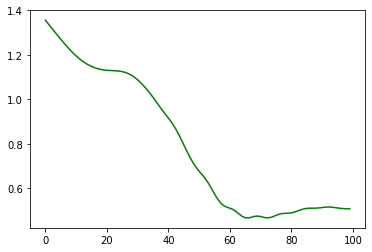
= ExactGPModel(train_x, train_y, gpytorch.likelihoods.GaussianLikelihood(), = gpytorch.ExactMarginalLogLikelihood(gp_model.likelihood, gp_model)def loss1(model, x, y):= torch.cholesky(model.likelihood(model(x)).covariance_matrix)= torch.cholesky_solve(y.reshape(- 1 ,1 ), l)return (y.reshape(- 1 ,1 ).T@ alp).ravel()def loss_det(model, x, y):= torch.cholesky(model.likelihood(model(x)).covariance_matrix)return torch.log(l.diagonal()).sum ()def loss_full(model, x, y):= model(x)return - zmll(dist, y)= torch.optim.Adam(gp_model.parameters(), lr= 0.1 )0 )for p in gp_model.parameters():= []= []= []for i in range (100 ):True )print (i)= 0 = zloss + loss1(gp_model, train_x, train_y)= zloss + loss_det(gp_model, train_x, train_y)# for i in range(100): # clear_output(True) # print(i) # zopt.zero_grad() # zloss = 0 # zloss = zloss + loss_det(gp_model, train_x, train_y) # zloss = zloss + loss1(gp_model, train_x, train_y) # zloss.backward() # zopt.step() # zlosses.append(loss2(gp_model, train_x, train_y).item()) = 'inverse term' ); = 'log det term' ); = 'full loss' ); ; = 'g' );
eval ()with torch.no_grad(), gpytorch.settings.fast_pred_var():= gp_model.likelihood(gp_model(test_x)); - 2 ,2 )