import jax
import jax.numpy as jnp
import numpy as np
from sklearn.datasets import make_blobs
import matplotlib.pyplot as plt
from matplotlib.animation import FuncAnimation
from time import time
# Enable high precision
from jax.config import config
config.update("jax_enable_x64", True)
# To enable animation inside notebook
plt.rc("animation", html="jshtml")Create dataset
features, labels = make_blobs(100, n_features=2, centers=2, random_state=0)
plt.scatter(features[:, 0], features[:, 1], c=labels);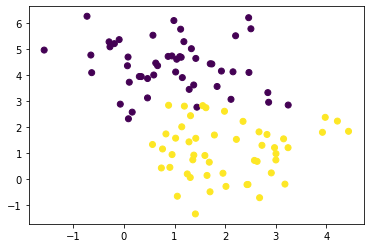
print(features.shape, features.dtype, labels.shape, labels.dtype)(100, 2) float64 (100,) int64Implementing Newton’s method (naive way)
We will first try to implement Eq. 10.31 directly from PML book1:
\[ \boldsymbol{w}_{t+1}=\boldsymbol{w}_{t}-\eta_{t} \mathbf{H}_{t}^{-1} \boldsymbol{g}_{t} \]
def get_logits(params, feature): # for a single data-point
logits = jnp.sum(feature * params["w"]) + params["b"]
return logits
def naive_loss(params, feature, label): # for a single data-point
logits = get_logits(params, feature)
prob = jax.nn.sigmoid(logits)
# Check if label is 1 or 0
is_one = (label == 1)
loss_if_one = lambda: -jnp.log(prob) # loss if label is 1
loss_if_zero = lambda: -jnp.log(1 - prob) # loss if labels is 0
# Use lax.cond to convert if..else.. in jittable format
loss = jax.lax.cond(is_one, loss_if_one, loss_if_zero)
return loss
def naive_loss_batch(params, features, labels): # for a batch of data-points
losses = jax.vmap(naive_loss, in_axes=(None, 0, 0))(params, features, labels)
return jnp.mean(losses)Writing the train function
def naive_train_step(params, features, labels, learning_rate):
# Find gradient
loss_value, grads = jax.value_and_grad(naive_loss_batch)(params, features, labels)
# Find Hessian
hess = jax.hessian(naive_loss_batch)(params, features, labels)
# Adjust Hessian matrix nicely
hess_matrix = jnp.block([[hess["b"]["b"], hess["b"]["w"]],
[hess["w"]["b"], hess["w"]["w"]]])
# Adjust gradient vector nicely
grad_vector = jnp.r_[grads["b"], grads["w"]]
# Find H^-1g
h_inv_g = jnp.dot(jnp.linalg.inv(hess_matrix), grad_vector)
# Get back the structure
h_inv_g = {"b": h_inv_g[0], "w": h_inv_g[1:]}
# Apply the update
params = jax.tree_map(lambda p, g: p - learning_rate*g, params, h_inv_g)
return params, loss_value
# First order method
# vg = jax.value_and_grad(naive_loss_batch)
# def train_step(params, features, labels, learning_rate):
# # Find gradient
# loss_value, grads = vg(params, features, labels)
# # Apply the update
# params = jax.tree_map(lambda p, g: p - learning_rate*g, params, grads)
# return params, loss_valuekey = jax.random.PRNGKey(0)
random_params = jax.random.normal(key, shape=(3, ))
# "b" should have shape (1,) for hessian trick with jnp.block to work
params = {"w": random_params[:2], "b": random_params[2].reshape(1,)}
learning_rate = 1.0
epochs = 20
train_step_jitted = jax.jit(naive_train_step)
history = {"loss": [], "params": []}
# warm up
train_step_jitted(params, features, labels, learning_rate)
init = time()
for _ in range(epochs):
history["params"].append(params)
params, loss_value = train_step_jitted(params, features, labels, learning_rate)
history["loss"].append(loss_value)
print(time() - init, "seconds")
print(params)WARNING:absl:No GPU/TPU found, falling back to CPU. (Set TF_CPP_MIN_LOG_LEVEL=0 and rerun for more info.)0.0015490055084228516 seconds
{'b': DeviceArray([13.22076694], dtype=float64), 'w': DeviceArray([ 0.59021174, -5.18797851], dtype=float64)}A helper function to animate the learning.
def animate(history):
fig, ax = plt.subplots(1, 2, figsize=(10,4))
def update(idx):
# Clear previous frame
ax[0].cla()
ax[1].cla()
# Plot data
params = history["params"][idx]
losses = history["loss"][:idx]
ax[0].scatter(features[:, 0], features[:, 1], c=labels)
# Calculate and plot decision boundary
x0_min, x0_max = features[:, 0].min(), features[:, 0].max()
x1_min = -(params["b"] + params["w"][0] * x0_min)/params["w"][1]
x1_max = -(params["b"] + params["w"][0] * x0_max)/params["w"][1]
ax[0].plot([x0_min, x0_max], [x1_min, x1_max], label='decision boundary')
# Plot losses
ax[1].plot(losses, label="loss")
ax[1].set_xlabel("Iterations")
ax[0].legend()
ax[1].legend()
anim = FuncAnimation(fig, update, range(epochs))
plt.close()
return animanimate(history)Implementing IRLS algorithm
def get_s_and_z(params, feature, label): # for a single data-point
logits = get_logits(params, feature)
prob = jax.nn.sigmoid(logits)
s = prob * (1 - prob)
z = logits + (label - prob)/s
return s, z
def irls_train_step(params, features, labels):
s, z = jax.vmap(get_s_and_z, in_axes=(None, 0, 0))(params, features, labels)
S = jnp.diag(s.flatten()) # convert into a diagonal matrix
# Add column with ones
X = jnp.c_[jnp.ones(len(z)), features]
# Get weights
weights = jnp.linalg.inv(X.T@S@X)@X.T@S@z.flatten()
# get correct format
params = {"b": weights[0], "w": weights[1:]}
return paramskey = jax.random.PRNGKey(0)
random_params = jax.random.normal(key, shape=(3,))
params = {"w": random_params[:2], "b": random_params[2]}
epochs = 20
train_step_jitted = jax.jit(irls_train_step)
irls_history = {"params": []}
# warm up
train_step_jitted(params, features, labels)
init = time()
for _ in range(epochs):
irls_history["params"].append(params)
params = train_step_jitted(params, features, labels)
print(time() - init, "seconds")
print(params)0.0016303062438964844 seconds
{'b': DeviceArray(13.22076694, dtype=float64), 'w': DeviceArray([ 0.59021174, -5.18797851], dtype=float64)}Comparison
naive_params_b = list(map(lambda x: x["b"], history["params"]))
irls_params_b = list(map(lambda x: x["b"], irls_history["params"]))
naive_params_w = list(map(lambda x: x["w"], history["params"]))
irls_params_w = list(map(lambda x: x["w"], irls_history["params"]))plt.plot(naive_params_b, "o-", label="Naive")
plt.plot(irls_params_b, label="IRLS")
plt.xlabel("Iterations")
plt.title("Bias")
plt.legend();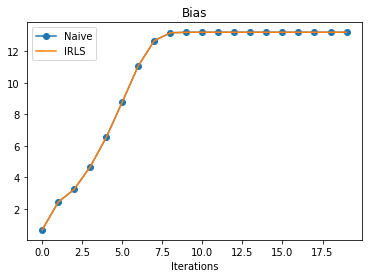
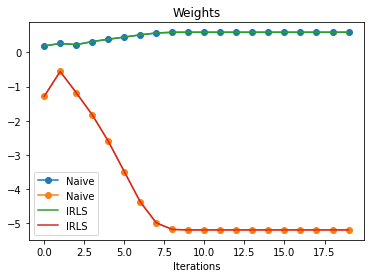
plt.plot(naive_params_w, "o-", label="Naive")
plt.plot(irls_params_w, label="IRLS")
plt.xlabel("Iterations")
plt.title("Weights")
plt.legend();